Researchers at Tsinghua University have developed a multi-agent system called OpenLens AI that can run an entire health-informatics study—from literature search to a finished LaTeX manuscript—without human help. The framework strings together separate agents for reviewing medical papers, analysing clinical datasets, writing code, and drafting publication-ready documents, all under a central supervisor. In internal tests on two intensive-care databases, the system delivered manuscripts that independent language-model judges rated as largely sound for routine statistical questions and moderately reliable for more complex tasks. The team says the project, available at openlens.icu, is the first end-to-end platform to combine vision-language feedback with domain-specific quality checks aimed at reducing medical-research errors.
Why health informatics needs automation
Clinicians and data scientists work with a wide mix of information—vital-sign streams, lab results, imaging and fast-growing literature. Sifting and combining that material is labour-intensive, and mistakes can have direct clinical consequences. Recent large-language-model (LLM) agents can already summarise papers or draft code, but they often miss two points that matter in medicine: understanding charts and medical images, and enforcing rigorous validation steps that catch data leakage or bad statistics.
OpenLens AI is designed to fill those gaps. It uses a vision-language model to inspect every figure it produces, from bar charts to causal diagrams, and includes checklists that verify citations, trace code to results, and flag common methodological pitfalls. The end product is a complete LaTeX manuscript formatted for journal submission, with links back to every script and dataset the agents touched.
Inside the five-agent workflow
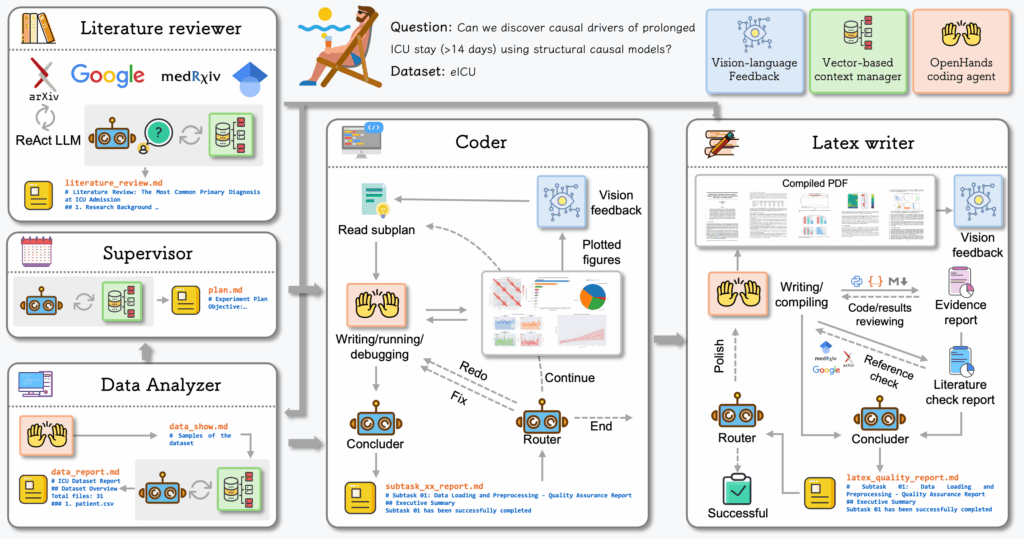
Instead of one large model doing everything, OpenLens AI splits the work across five specialised agents that share a persistent “state” file recording every intermediate result.
- Supervisor. Breaks a user’s one-line idea into ordered subtasks and hands them to the right agent.
- Literature reviewer. Searches arXiv, MedRxiv and other repositories, then writes a concise review with ReAct-style tool use. All references go through a citation check to weed out fabricated papers.
- Data analyzer. Turns raw clinical tables or time-series into cleaned datasets, visualisations and narrative summaries. A router decides whether results need more processing or can move on.
- Coder. Generates and runs Python scripts via the OpenHands toolkit, rerunning or fixing code when execution or vision checks fail. A vision-language model scores every figure for clarity and correctness.
- LaTeX writer. Collects approved figures, drafts each manuscript section, and loops through polish cycles until either a quality threshold or an iteration cap is hit.
All agents are orchestrated with LangGraph, which forces workflows to follow a directed graph and allows automatic retries if a node crashes. Every claim in the final PDF links back to the exact SQL query or Python cell that produced it, giving reviewers a complete audit trail.
Benchmarking on two ICU datasets
Because no standard test bed exists for autonomous medical-research agents, the team built an 18-task benchmark covering easy, medium and hard questions. Tasks ranged from simple counts (e.g. distribution of patient ages) to causal modelling of prolonged ICU stays. The experiments used public subsets of the MIMIC-IV and eICU databases so that results can be reproduced.
To keep the system deployable in hospitals that prefer on-premise hardware, the authors opted for mid-size models: GLM-4.5-Air as the language backbone and GLM-4.1V-9B-Thinking for vision-language checks. Each task allowed at most two cycles of code-refinement and two passes of LaTeX polishing.
Performance was scored by a separate language model acting as a “judge” across five criteria: plan completion, code execution, result validity, paper completeness and conclusion quality. Scores run from 1 (severe issues) to 3 (minor or none). Averages across the nine publicly documented tasks are shown below.
| Difficulty |
Example task |
Average score
(1 = poor, 3 = good) |
| Easy |
Age distribution by gender |
≈ 2.8 |
| Medium |
Predict 30-day mortality |
≈ 2.4 |
| Hard |
Causal drivers of long ICU stay |
≈ 2.1 |
For straightforward descriptive analytics, the system earned near-perfect marks, producing clean code and sound conclusions. Medium tasks saw occasional missteps in preprocessing or model fitting, though results still reached “moderate” quality. The hardest causal and generalisation questions exposed current limits: the agent sometimes picked inappropriate methods or missed confounders, prompting lower validity scores.
How to Install OpenLens Ai
# Clone the repository
git clone [email protected]:jarrycyx/openlens-ai.git
cd agent-med
# Install dependencies
conda create -n py312 python=3.12
conda activate py312
pip install --upgrade pip
pip install -e .
# Configure environment variables
cp .env.example .env
# Edit .env with your API keys and model settings
Why it Matters Now?
Medical AI has surged ahead in diagnosis and imaging, but turning raw hospital data into peer-reviewable research still takes months and teams of statisticians. OpenLens AI shows that large parts of that workflow can now run on autopilot, at least for routine descriptive studies. A clinician could drop a single-line question—say, “How many ICU patients with pneumonia die in-hospital?”—and receive a polished manuscript complete with vetted figures and citations.
That speed could help hospitals extract value from the vast troves of electronic-health-record data they already collect, especially in smaller centres without in-house data scientists. The built-in audit trail is also a step toward making AI-generated research more trustworthy, an issue that has dogged earlier text-generation tools.
Open questions and next steps
The authors acknowledge several limits. First, there is no head-to-head comparison with general-purpose agents, largely because no common benchmark exists. Second, mid-size models may struggle with highly complex or multimodal data, such as raw imaging or genomics. Third, the evaluation covered only a slice of MIMIC-IV and eICU; broader datasets and real-world hospital deployments will be needed to test generalisability.
Future work will focus on larger public benchmarks, fine-tuning both language and vision modules for medical jargon, and adding privacy features for on-premise use. The team is also exploring more advanced causal-inference agents to boost performance on the hardest tasks.
For now, OpenLens AI is available under an open-source licence on GitHub, letting others inspect the code and replicate the benchmark results. Whether autonomous agents can handle the full rigor of clinical research is still up for debate, but Tsinghua’s work suggests the gap is narrowing fast.
You can explore more on OpenLens Ai here at their website – openlens.icu and the full paper.